A predictive model has been developed that enables researchers to encode instructions for cells to execute.
Scientists at the University of California, San Francisco (UCSF) and IBM Research have created a virtual library of thousands of “command sentences” for cells using machine learning. These “sentences” are based on combinations of “words” that direct engineered immune cells to find and continuously eliminate cancer cells.
This research, which was recently published in the journal Science, is the first time that advanced computational techniques have been applied to a field that has traditionally progressed through trial-and-error experimentation and the use of pre-existing molecules rather than synthetic ones to engineer cells.
The advance allows scientists to predict which elements – natural or synthesized – they should include in a cell to give it the precise behaviors required to respond effectively to complex diseases.
“This is a vital shift for the field,” said Wendell Lim, Ph.D., the Byers Distinguished Professor of Cellular and Molecular Pharmacology, who directs the UCSF Cell Design Institute and led the study. “Only by having that power of prediction can we get to a place where we can rapidly design new cellular therapies that carry out the desired activities.”
Meet the Molecular Words That Make Cellular Command Sentences
Much of therapeutic cell engineering involves choosing or creating receptors that, when added to the cell, will enable it to carry out a new function. Receptors are molecules that bridge the cell membrane to sense the outside environment and provide the cell with instructions on how to respond to environmental conditions.
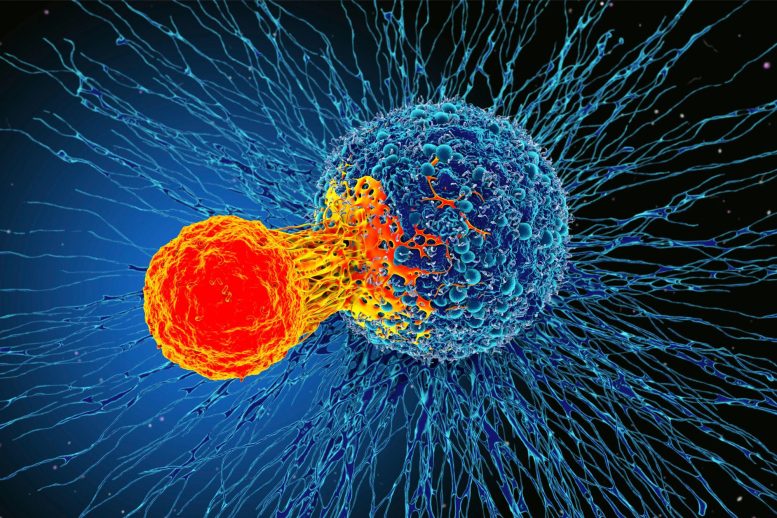
Putting the right receptor into a type of immune cell called a T cell can reprogram it to recognize and kill cancer cells. These so-called chimeric antigen receptors (CARs) have been effective against some cancers but not others.
Lim and lead author Kyle Daniels, Ph.D., a researcher in Lim’s lab, focused on the part of a receptor located inside the cell, containing strings of amino acids, referred to as motifs. Each motif acts as a command “word,” directing an action inside the cell. How these words are strung together into a “sentence” determines what commands the cell will execute.
Many of today’s CAR-T cells are engineered with receptors instructing them to kill cancer, but also to take a break after a short time, akin to saying, “Knock out some rogue cells and then take a breather.” As a result, the cancers can continue growing.
The team believed that by combining these “words” in different ways, they could generate a receptor that would enable the CAR-T cells to finish the job without taking a break. They made a library of nearly 2,400 randomly combined command sentences and tested hundreds of them in T cells to see how effective they were at striking leukemia.
What the Grammar of Cellular Commands Can Reveal About Treating Disease
Next, Daniels partnered with computational biologist Simone Bianco, Ph.D., a research manager at IBM Almaden Research Center at the time of the study and now Director of Computational Biology at Altos Labs. Bianco and his team, researchers Sara Capponi, Ph.D., also at IBM Almeden, and Shangying Wang, Ph.D., who was then a postdoc at IBM and is now at Altos Labs, applied novel machine learning methods to the data to generate entirely new receptor sentences that they predicted would be more effective.
“We changed some of the words of the sentence and gave it a new meaning,” said Daniels. “We predictively designed T cells that killed cancer without taking a break because the new sentence told them, ‘Knock those rogue tumor cells out, and keep at it.’”
Pairing machine learning with cellular engineering creates a synergistic new research paradigm.
“The whole is definitely greater than the sum of its parts,” Bianco said. “It allows us to get a clearer picture of not only how to design cell therapies, but to better understand the rules underlying life itself and how living things do what they do.”
Given the success of the work, added Capponi, “We will extend this approach to a diverse set of experimental data and hopefully redefine T-cell design.”
The researchers believe this approach will yield cell therapies for autoimmunity, regenerative medicine, and other applications. Daniels is interested in designing self-renewing stem cells to eliminate the need for donated blood.
He said the real power of the computational approach extends beyond making command sentences, to understanding the grammar of the molecular instructions.
“That is the key to making cell therapies that do exactly what we want them to do,” Daniels said. “This approach facilitates the leap from understanding the science to engineering its real-life application.”
- aum
-

 1
1



Recommended Comments
There are no comments to display.
Join the conversation
You can post now and register later. If you have an account, sign in now to post with your account.
Note: Your post will require moderator approval before it will be visible.