Mutations are the raw ingredient of evolution, providing variation that sometimes makes an organism more successful in its environment. But most mutations are expected to be neutral and have no impact on an organism's fitness. These can be incredibly useful since these incidental changes help us track evolutionary relationships without worrying about selection for or against the mutation affecting its frequency. All of the genetic ancestry tests, for example, rely heavily on tracking the presence of these neutral mutations.
But this week, a paper provided evidence that a significant category of mutations isn't as neutral as we thought they were. The big caveat is that the study was done in yeast, which is a weird organism in a couple of ways, so we'll have to see if the results hold in others.
True neutral?
One of the reasons that most mutations are neutral is that most of our DNA doesn't seem to be doing anything useful. Only a few percent of the human genome is composed of the portion of genes that encode proteins, and only some of the nearby DNA is involved in controlling the activity of those genes. Outside of those regions, mutations don't do much, either because the DNA there has no function or because the function isn't very sensitive to having a precise sequence of bases in the DNA.
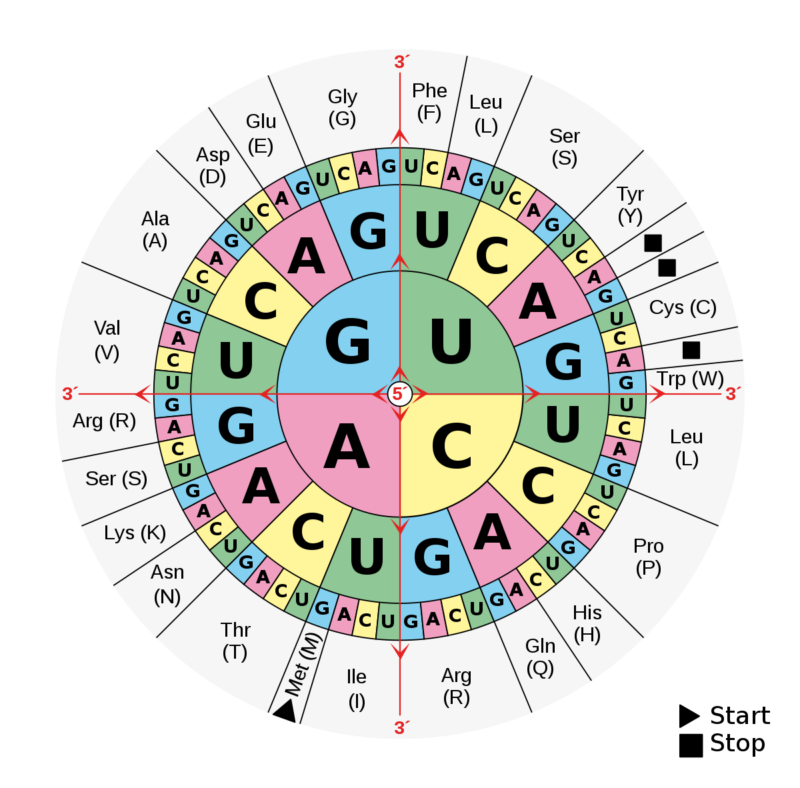
But even within the parts of genes that encode proteins, the precise sequence shouldn't matter all that much. Each protein's amino acid is encoded by a combination of three bases in DNA. That means there are 64 possible codes for amino acids—but we only use 20 different amino acids. As a result, there's plenty of redundancy in the genetic code. For example, the base series ACG encode the amino acid threonine. So does the series ACA. And ACC. All told, four different codes will get you threonine.
The key thing to note is that all four codes start with AC. If you have a mutation in either of those two bases, you no longer get threonine. But if you get a mutation in the third position, it doesn't matter—whatever you change the base to, you still get threonine. That should be a completely neutral mutation. And researchers have used the assumption that it is neutral to help them track protein evolution.
That's the assumption that the new paper put to the test.
Make all the mutations
To test neutral mutations, the researchers started with a panel of 21 yeast genes, chosen partly because they are involved in a wide variety of cellular activities. The other part behind their choice is that eliminating these genes doesn't kill the yeast but makes it less healthy. That should make it easier to detect partial effects, where the mutation makes the yeast less healthy.
Within that stretch, the researchers picked a 150-base stretch in the DNA and made every single possible mutation, using DNA editing to make a yeast strain carrying the mutation. That is a total of over 9,000 individual yeast strains, with some carrying mutations that will change the amino acid sequence and others carrying mutations we'd expect to be neutral. But of course, this involved lab work, where things don't work for random, unknown reasons, so the researchers had to settle for testing about 8,300 mutant yeast strains.
The test was pretty simple. Throw equal numbers of normal and mutant yeast in a flask, and let them grow for a bit. Then, sample the population, and check the relative levels of normal and mutant yeast. If the mutation lowered the fitness, you'd see more normal yeast when you sampled the flask.
That was true for mutations that changed an amino acid. These saw their relative fitness drop a bit, though not by much (their fitness was 0.988 that of the normal yeast). But the neutral mutations weren't notably different—they also dropped the yeast's fitness by a tiny amount relative to a normal strain. In effect, the mutations that didn't change any amino acids were, on average, indistinguishable from the ones that did. Beyond this average, you could tell a slight difference. There were more amino acid-altering mutations that had a stronger deleterious effect on fitness, and more neutral ones that had a minimal effect. But it's clear that, as a whole, the class expected to be neutral wasn't.
Wait, what?
On the surface, that doesn't make any sense. Both versions of the gene encode exactly the same amino acid. How could one possibly be less fit than the other?
The secret to understanding this is remembering that the gene's DNA isn't used directly to make a protein. Instead, it's transcribed into an RNA copy called a messenger RNA, and that is directly translated to make the protein. And alterations in the DNA can affect the RNA's three-dimensional structure, its stability in the cell, and the rate it's translated into protein.
The researchers found that the mutations expected to be neutral often influenced the amount of messenger RNA present in the cell. They also appeared to influence the RNA's ability to fold into a three-dimensional shape. And they were also likely to affect the efficiency of the messenger RNA in translating into a protein. Combined, these could account for why this group of mutations had a collective impact on fitness.
So, does that mean it's time to throw out our idea that mutations in a gene that don't alter its protein sequence are neutral? And with it, all the tools we use to study protein evolution that are based on this assumption?
The researchers give one major reason why this would be premature: Yeasts are kind of weird. To start, unlike animals, which mostly get a copy of a gene from their moms and another from their dads, yeast carries only one copy of every gene, so it will likely be more sensitive to subtle effects. Yeasts also live a lifestyle similar to bacteria, carrying a simplified genome and focusing on rapid reproduction—a relatively minor metabolic hit is more likely to slow them down.
And these effects are still very subtle. Even if you completely hammered the function of these proteins by creating a mutation that truncated the protein early, the fitness cost was slight (fitness was 0.94 of the yeast strain without any mutations). It's not even clear this behavior would occur in other genes in yeast, much less genes in other organisms.
The other thing that the researchers note is that, in actual populations that are evolving over the long term, evolutionary pressures are constantly shifting due to environmental changes. So, it could be that these mutations are effectively neutral in a realistic environment, so when we look at similar mutations in a natural population, they appear to be neutral.
All these results are an important caution and make it worth the time and effort needed to sort this out carefully.
Nature, 2022. DOI: 10.1038/s41586-022-04823-w (About DOIs).


3175x175(CURRENT).thumb.jpg.b05acc060982b36f5891ba728e6d953c.jpg)

Recommended Comments
There are no comments to display.
Join the conversation
You can post now and register later. If you have an account, sign in now to post with your account.
Note: Your post will require moderator approval before it will be visible.