MicroRNAs control the activity of many key genes but were unknown before 1993.
On Monday, the Nobel Committee announced that two US researchers, Victor Ambros and Gary Ruvkun, will receive the prize in Physiology or Medicine for their discovery of a previously unknown mechanism for controlling the activity of genes. They discovered the first of what is now known to be a large collection of MicroRNAs, short (21-23 bases long) RNAs that bind to and alter the behavior of protein-coding RNAs. While first discovered in a roundworm, they've since been discovered to play key roles in the development of most complex life.
The story behind the discovery is typical of a lot of the progress in the biological sciences: genetics helps identify a gene important for the development of one species, and then evolutionary conservation reveals its widespread significance.
In the worm
Ambros and Ruvkun started on the path to discovery while post-doctoral fellows in the lab of earlier Nobel winner Robert Horvitz, who won for his role in developing the roundworm C. elegans as an experimental genetic organism. As part of the early genetic screens, people had identified a variety of mutations that caused developmental problems for specific lineages of cells. These lin mutations included lin-4, which Ambros was characterizing. It lacked a number of specialized cell types, as well as the physical structures that depended on them.
Ruvkun was working on lin-14. The protein encoded by this gene is normally active early in development, but mutations that allowed it to persist to later stages cause early embryonic cell types to continue to develop. As they moved on to faculty positions, Ambros and Ruvkun both made some key discoveries about the system. Ambros demonstrated that lin-4 acts to block the activity of lin-14, while Ruvkun found that the regulation occurred after the lin-14 gene produced a mature messenger RNA. (A lot of gene regulation focuses on determining whether or not a messenger RNA is produced in the first place.) Some of the mutations that altered the activity of lin-14 turned out to be in a part of the RNA that doesn't code for a protein (the 3' untranslated region).
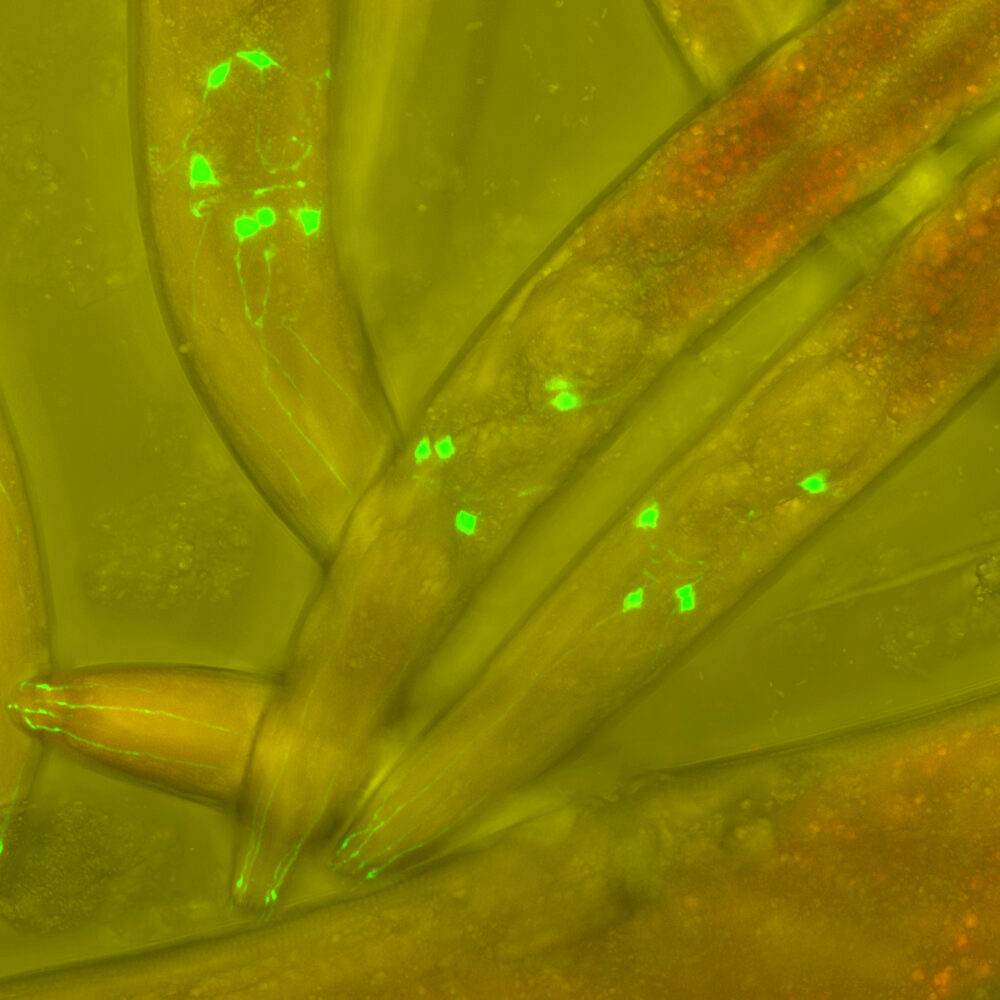
The key to the discovery took place in Ambros' lab, which narrowed down the DNA that contained lin-4 to a small region of DNA that didn't appear to include any protein-coding genes. Eventually, he was able to demonstrate that the area the gene was in encoded two extremely short RNAs: a 61 base long one, and one that was just a 22 bases subset of the longer one. At that point, Ambros and Ruvkun exchanged the sequences of their genes, and both recognized that the lin-4 RNA could base-pair with the portion of the lin-14 RNA that was critical to its regulation.
And that's where things stayed for the better part of a decade. With just one example of a microRNA from a single species that was notable for a number of developmental oddities, there was little indication that microRNAs were a significant biological phenomenon. But Ruvkun had ended up characterizing a gene called let-7 (let stands for lethal—animals die when eggs build up and explode through the animal's side) that also turned out to be a microRNA.
Critically, sequences related to let-7 show up in a huge range of animals, including other experimental organisms, like zebrafish and fruit flies, but also more distant relatives, like mollusks and humans.
And everywhere else
With their significance established, biochemical characterization of microRNAs has identified how the system operates. The longer form of the RNA produced from a microRNA gene acts as a precursor. It's able to fold over and base-pair with itself, forming a structure called a hairpin. An enzyme called Dicer cleaves it, forming the shorter, mature microRNA, which is able to base-pair with messenger RNAs. These mature microRNAs recruit a complex of proteins to the messenger RNA, which either causes the RNA to be digested or prevents them from being translated into proteins.
Based on the stereotypical hairpin structure, researchers have scanned genomes and found over 38,000 likely precursors; nearly 50,000 mature microRNAs have been discovered by sequencing all the RNA found in cells from a variety of species. While found widely in animals, they've also been discovered in plants, raising the possibility that they existed in a single-celled ancestral organism.
While some microRNA genes, including lin-4 and let-7, have dramatic phenotypes when mutated, many have weak or confusing effects. This is likely in part due to the fact that a single microRNA can bind to and regulate a variety of genes and so may have a mix of effects when mutated. In other cases, several different microRNAs may bind to the same messenger RNA, creating a redundancy that makes the loss of a single microRNA difficult to detect.
Nevertheless, there's plenty of evidence that, collectively, they're essential for normal development in many organisms and tissues. Knocking out the gene that encodes the Dicer protein, which is needed for forming mature microRNAs, causes early embryonic lethality. Knockouts of the gene in specific cell types cause a variety of defects. For example, B cells never mature if Dicer is lost in that cell lineage, and a knockout in nerve cells causes microcephaly and limiting branching of connections among neurons, leading the animals to die shortly after birth.
This being the Medicine prize, the Nobel Committee also cite a number of human genetic diseases that are caused by mutations in microRNA genes.
Overall, the award highlights just how complex life is at the cellular level. There's a fair number of genes that have to be made by every cell simply to enable their survival. But as for the rest, they exist embedded in complex regulatory networks that interact to ensure that proteins are made only where and when they're needed, and often degraded if they somehow get made anyway. And every now and then, fundamental research in an oddball species is still telling us unexpected things about those networks.
RIP Matrix | Farewell my friend ![]()
Hope you enjoyed this news post.
Thank you for appreciating my time and effort posting news every day for many years.
2023: Over 5,800 news posts | 2024 (till end of September): 4,292 news posts
- DKT27
-

 1
1


3175x175(CURRENT).thumb.jpg.b05acc060982b36f5891ba728e6d953c.jpg)
Recommended Comments
There are no comments to display.
Join the conversation
You can post now and register later. If you have an account, sign in now to post with your account.
Note: Your post will require moderator approval before it will be visible.