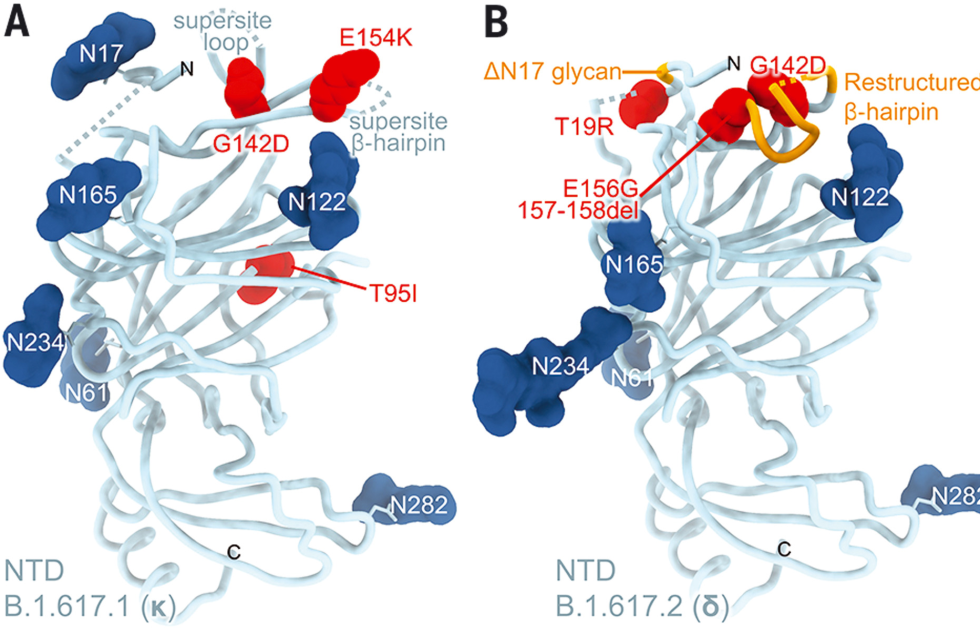
Right from omicron's first description, researchers were concerned about its variant of the SARS-CoV-2 virus. Looking over the list of mutations it carried, scientists could identify a number that would likely make the variant more infectious. Other mutations were even more worrying, as they would likely interfere with the immune system's ability to recognize the virus, allowing it to pose a risk to those who had been vaccinated or suffered from previous infections.
Buried in the subtext of these worries was a clear implication: scientists could simply look at the sequence of amino acids in the spike protein of a coronavirus and get a sense of how well the immune system would respond to it.
That knowledge is based on years of studying how the immune system operates, combined with a lot of specific information regarding its interactions with SARS-CoV-2. What follows is a description of these interactions, along with their implications for viral evolution and present and future variants.
Ts and Bs
To understand the immune system's function, it's easiest to break its responses into categories. To begin with, there's the innate immune response, which tends to recognize general features of pathogens rather than specific properties of individual bacteria or viruses. The innate response doesn't get fine-tuned by vaccination or prior exposure to a virus, so it's not really relevant to the discussion of variants.
What we're interested in is the adaptive immune response, which recognizes specific features in pathogens and generates a memory that produces a rapid and robust response if the same pathogen is ever seen again. It's the adaptive immune response that we're stimulating with vaccines.
The adaptive response can also be broken into categories. In terms of the relevant immune responses, we care most about those mediated by antibody-producing B cells. The other major part of adaptive immunity, the T cell, uses a completely different mechanism for identifying pathogens. We know a lot less about the T cell response to SARS-CoV-2, but we'll come back to that later. For now, we'll focus on antibodies.
Antibodies are large (in molecular terms) assemblies of four proteins. Most of the proteins are the same in all antibodies, which allows immune cells to interact with them. But each of the four proteins has a variable region that is different in every B cell produced. Many of the variable regions are useless, and others recognize the body's own proteins and get eliminated. But by chance, some antibodies have variable regions that recognize a segment of a protein made by a pathogen.
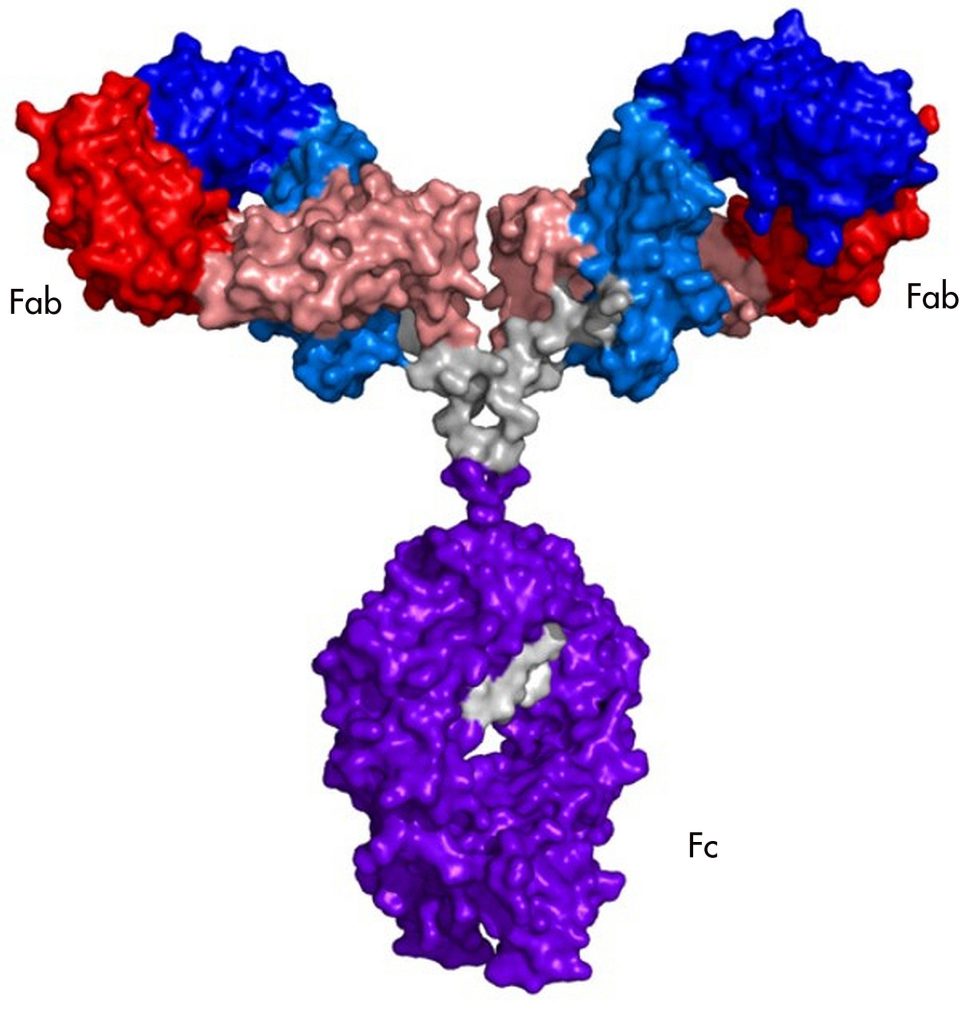
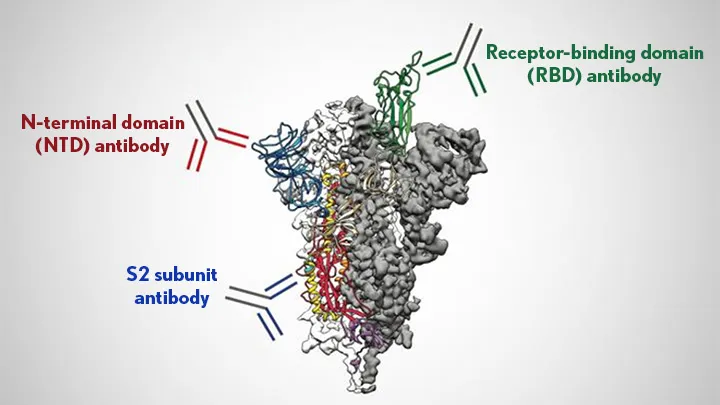
The portion of the pathogen's protein that the antibody recognizes is called an epitope. Epitopes vary from protein to protein, but they share some features. They have to be on the exterior of the protein, rather than buried in its interior, for the antibody to bump into it in the first place. And they often have amino acids that are polar or have a charge, since these form stronger interactions with the antibody.
You can't simply look at the amino acids in an antibody and tell what it's going to bind to. But if you have sufficient quantities of a specific antibody, it's possible to do what's called "epitope mapping," which involves figuring out precisely where on a protein the antibody is binding. In some cases, this can include a precise list of the amino acids that the antibody recognizes.
In general, having antibodies stuck to a pathogen in the bloodstream makes it easier for the pathogen to be spotted and disposed of by specialized immune cells—for this function, it really doesn't matter where the antibody sticks. But there are also specific interactions that can inactivate a virus in some cases, as we'll see below.


3175x175(CURRENT).thumb.jpg.b05acc060982b36f5891ba728e6d953c.jpg)

Recommended Comments
There are no comments to display.
Join the conversation
You can post now and register later. If you have an account, sign in now to post with your account.
Note: Your post will require moderator approval before it will be visible.