For the first time, scientists have sequenced the genome of a mysterious species of giant bacterium that can be seen without a microscope.
The discoveries about their reproductive strategies, survival mechanisms, and distinct metabolic mechanisms – similar to mitochondria – may one day be useful in developing sustainable energy technologies and increasing efficiency in agriculture.
Epulopiscium bacteria live symbiotically in the guts of a fish, Naso tonganus, in tropical ocean environments. Where most bacteria are too small to be seen without a microscope, these single-celled mammoths have a million times the volume of their well-known relatives, E. coli, meaning they can just be made out with the naked eye.
"This incredible giant bacterium is unique and interesting in so many ways," says microbiologist Esther Angert from Cornell University in the US. "Revealing the genomic potential of this organism just kind of blew our minds."
The first member of Epulopiscium – whose name comes from the Latin words for "a guest" and "of a fish" – was discovered in 1985.
Angert and her US colleagues named the species they studied Epulopiscium viviparus: The second word refers to reproduction that results in live births.
While bacteria typically split in half to make two new ones, E. viviparus can make up to 12 copies of itself inside the parent cell, which then swim out into the world.
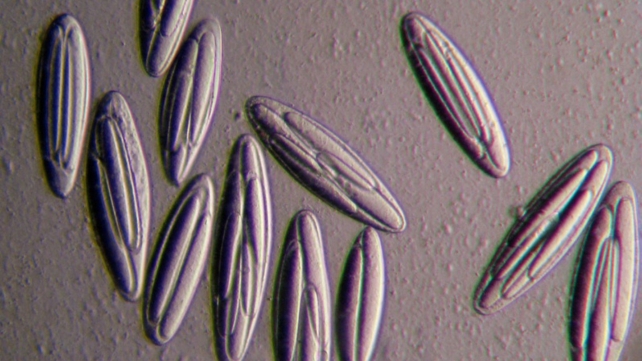
Micrograph of a group of Epulopiscium viviparus bacteria. (Esther Angert)
Unable to be grown in a lab, such giant bacteria remain a curiosity of the biological world. So to study E. viviparus, the researchers had to capture the fish it lives in and carefully collect cells as quickly as possible for DNA sequencing and transcriptome analysis.
Most bacteria either respire using oxygen or obtain energy from their environment through fermentation, which generally results in less energy production.
E. viviparus happens to be a fermenter, but that's puzzling because it's huge, reproduces rapidly, and can swim – all requiring relatively large amounts of energy.
It looks like the bacteria have optimized their metabolism to a fish gut environment that's rich in sodium ions. The flow of sodium ions across cell membranes generates a powerful 'sodium motive force' for making energy and spinning their hairlike appendages called flagella for movement.
This sodium motive force also powers flagella movement in Vibrio cholerae, the bacteria that cause cholera.
The team also found a large chunk of the genetic code of E. viviparus makes enzymes that are highly efficient at nutrient extraction from their host fish, especially carbohydrates called polysaccharides from the algae that forms a large part of N. tonganus's diet.
E. viviparus has plentiful enzymes that make ATP too, the 'energy currency' that supports a wide variety of cellular processes. They discovered space for these molecules in a unique membrane, similar to the mitochondria of more complex organisms.
"We all know that phrase "the mitochondria are the powerhouse of the cell", and amazingly, these membranes in E. viviparus have kind of converged on the same model as the mitochondria," Angert says.
"They have a highly folded membrane that increases surface area where these energy-producing pumps can work, and that increased surface area creates a powerhouse of energy."
E. viviparus's efficient ways of harnessing the nutrients in algae could have a lot of uses in the future. Algae is becoming more popular as a source of renewable energy, a food source for livestock, and for people too, because its growth doesn't interfere with land-based farming.
There are still some mysteries to be solved. As Angert and colleagues point out, further study is required to completely understand how E. viviparus uses its arsenal of enzymes. But this provides a solid foundation for understanding their growth requirements.
"We find it remarkable that the largest known bacteria have thus far eluded isolation," the authors write. "This suggests that bacterial behemoths are highly tuned to survive in the environments in which they evolved."
The study has been published in Proceedings of the National Academy of Sciences.
- Adenman
-

 1
1


Recommended Comments
There are no comments to display.
Join the conversation
You can post now and register later. If you have an account, sign in now to post with your account.
Note: Your post will require moderator approval before it will be visible.