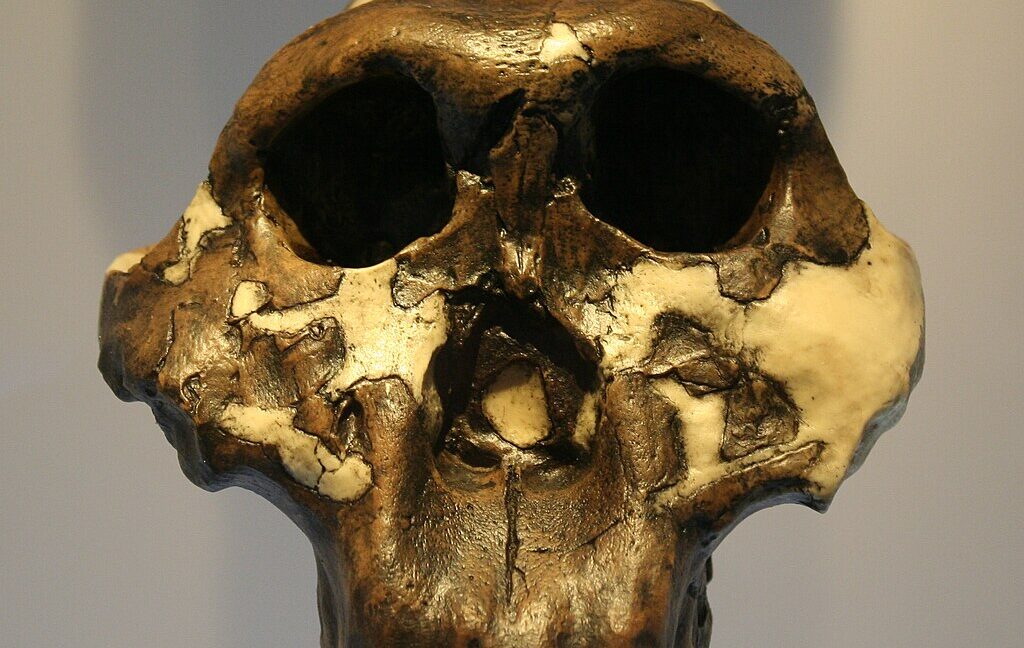
We weren't even sure if Paranthropus remains come from a single species.
The ability to study ancient DNA has revolutionized our ability to understand our own species' past. It has clarified our relationship with Neanderthals and revealed the existence of Denisovans. But even in the most favorable environments, DNA degrades over time, setting a limit on how far back we can hope to resolve questions about our ancestors. And most of the species we've had trouble understanding lived in Africa, where the conditions are far less favorable for DNA's survival.
But a large international team has now found another way to get some information about the genetics out of far older remains. They've extracted fragments of enamel proteins from the teeth of fossils of the species Paranthropus robustus and used them to test whether the remains truly belonged to one species, despite dramatic differences in size. Because one of the proteins is male-specific, they also found the size of the individual wasn't necessarily related to its sex.
A complicated species
Remains that have been classified as Paranthropus show up in the fossil record nearly 3 million years ago and persist for roughly a million years. That means it overlapped both with australopithecines and early members of the Homo genus. Four different species have been assigned to this genus, but the situation is complicated. It shares a lot of similarities with some species of Australopithecus, raising the possibility of interbreeding. There's also a lot of variation within remains identified as Paranthropus, notably in the size of individuals. Some have suggested that this might be due to male/female differences in this species (termed "sexual dimorphism"), but that has been difficult to test.
Unfortunately, no remains have been found outside of Africa, and no DNA older than 20,000 years has been recovered from that continent—well after Paranthropus went extinct. So, for Paranthropus remains that date from roughly 2 million years ago, the team turned to proteins found in teeth. These inevitably get damaged over time—broken down into smaller fragments, with some amino acids chemically altered. But the robust nature of tooth enamel should prevent their wholesale destruction.
And the technique known as mass spectrometry is sensitive enough to identify individual isotopes of atoms incorporated into larger molecules. Using this, the researchers could identify the likely composition of individual fragments of proteins and match those to known tooth enamel proteins.
After testing their techniques on animal remains found with the hominins, the researchers turned to samples from the Paranthropus teeth, all found at the same site within South Africa’s Cradle of Humankind World Heritage Site. While the proteins identified varied from sample to sample, they found six different proteins that were present in all four of the teeth studied, though again, only as fragments. Collectively, those fragments covered 425 amino acids of the six proteins.
To confirm that the results, generated in a lab in Copenhagen, were real, they repeated the process at a lab in Cape Town. The successful replication was bolstered by the fact that some of the amino acids showed signs of chemical damage, indicating that they really were pieces of 2 million-year-old proteins.
Reading amino acids
So, what could small bits of ancient proteins tell us? In this case, more than you might expect. That's because one of the enamel proteins, AMELY, comes from a gene that's located on the Y chromosome (there's also an AMELX gene on the X chromosome). And some of the protein fragments identified in a subset of the four remain clearly included pieces of AMELY, indicating that the tooth had come from a male individual.
That turns out to be critical to testing the idea that body size is an indication of sex differences in Paranthropus. One of the samples was a small tooth that had been suggested to be from a female individual, but the presence of AMELY unambiguously identifies it as male. That's an important result because, as the researchers put it, "it enables us to exclude sexual dimorphism as one of the multiple variables affecting the range of anatomical variation," in this species.
The absence of AMELY suggests that a sample is female, but it isn't definitive. That's both because it's impossible to rule out some problem with identifying the protein in samples this old, and in part because some rare males (including at least one Neanderthal) carry deletions that eliminate the gene entirely.
Another key aspect is that some of the 425 amino acid locations differ between hominin species, and even individual members of Paranthropus. Thus, they can potentially serve as a diagnostic of the relationships between and within species and help address some of the confusion about how many species of Paranthropus there were and their relationship with other hominins. While it's difficult to say too much with only four samples, the researchers found some suggestive evidence.
For example, they tested whether you might see the sort of amino acid variation found among these samples if they all belonged to the same species. This was done by randomly choosing four human genomes and examining whether they had a similar level of variation. They concluded that it was "plausible" that you'd see this level of variation among any four individuals that were chosen at random, but the population of modern humans is likely to be larger than that of Paranthropus, so the test wasn't definitive.
Among the 425 different amino acids were 16 that had species-specific variations among hominins. Somewhat surprisingly, Paranthropus robustus is the most closely related species to our own genus, Homo, based on a tree built from these variations. Again, however, they conclude that there simply isn't enough data available to feel confident in this conclusion.
But that should really be an "isn't enough data yet." We heard about this paper from regular Ars reader Enrico Cappellini, who happens to be its senior author and faculty at the University of Copenhagen's Globe Institute. And a quick look over his faculty profile indicates that developing the techniques used here is his major research focus, so hopefully we'll be able to expand the data available on extinct hominin species with time. The challenge, as noted in the paper, is that the technique destroys a small part of the sample, and these samples are one-of-a-kind pieces of the collective history of all of humanity.
Science, 2025. DOI: 10.1126/science.adt9539 (About DOIs).
Hope you enjoyed this news post.
Thank you for appreciating my time and effort posting news every day for many years.
News posts... 2023: 5,800+ | 2024: 5,700+ | 2025 (till end of April): 1,811
RIP Matrix | Farewell my friend ![]()


3175x175(CURRENT).thumb.jpg.b05acc060982b36f5891ba728e6d953c.jpg)
Recommended Comments
Join the conversation
You can post now and register later. If you have an account, sign in now to post with your account.
Note: Your post will require moderator approval before it will be visible.