Traditional antibiotics drive bacteria toward drug resistance, so scientists are looking to viruses, CRISPR, designer molecules and protein swords for better treatments.
The bacteria may have entered her flesh along with shrapnel from the bomb detonated in Brussels Airport in 2016. Or perhaps the microbes hitched a ride on the surgical instruments used to treat her wounds. Either way, the "superbug" refused to be vanquished, despite years of antibiotic treatment.
The woman had survived a terrorist attack but was held hostage by drug-resistant Klebsiella pneumoniae, a bacterial strain often picked up by surgery patients in hospitals. Only by combining antibiotics with a new, experimental treatment did doctors finally rid her of the superbug.
Devastating drug-resistant bacterial infections like this one are all too common, and they represent an ever-growing threat to global health. In 2019, antibiotic-resistant bacteria directly killed roughly 1.27 million people worldwide and contributed to an additional 3.68 million deaths. In the U.S. alone, drug-resistant bacteria and fungi together cause an estimated 2.8 million infections and 35,000 deaths each year.
And the problem is getting worse: Seven of the 18 concerning bacteria tracked by the Centers for Disease Control and Prevention (CDC) are becoming more resistant to common antibiotics considered essential for maintaining public health. Meanwhile, drug companies have been slow to make new antibiotics capable of beating the microbes. Fewer than 30 antibiotics currently in the development pipeline target "priority" bacteria, as defined by the World Health Organization (WHO), and most of those drugs are still vulnerable to resistance, just like their predecessors.
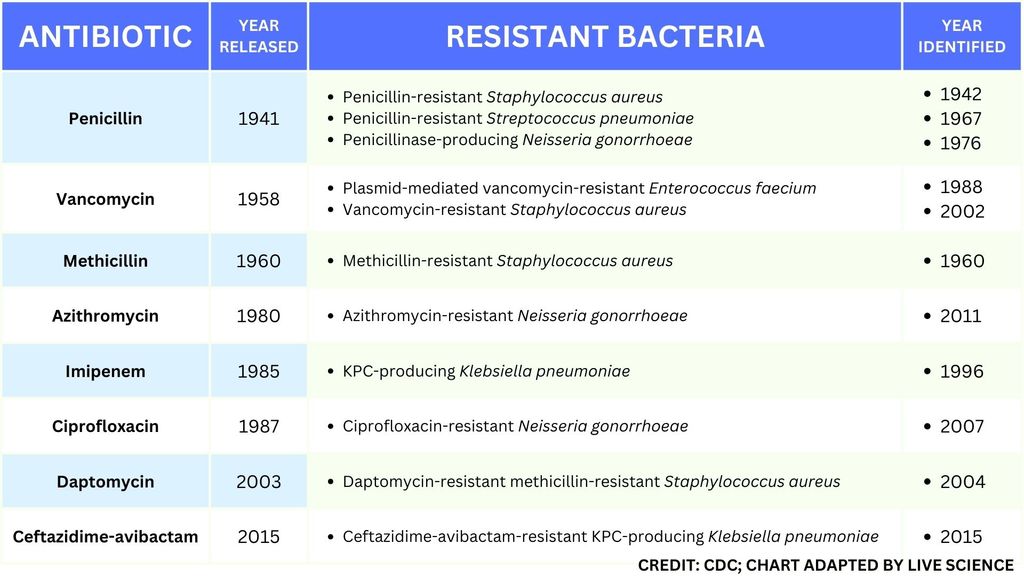
This table of select antibiotic-resistant bacteria demonstrates how rapidly important types of resistance developed after the approval and release of new antibiotics. (Image credit: Centers for Disease Control and Prevention. Adapted by Live Science from the CDC's "Select Germs Showing Resistance Over Time" Fact Sheet.)
So some scientists are looking beyond traditional antibiotics for new weapons that won't fuel the rise of superbugs. Their emerging arsenal features viruses that kill bacteria; CRISPR; and microbe-slaying molecules. They hope that these experimental treatments, some of which have been tested in patients, will kill superbugs without promoting resistance.
"The vision, for me, is that we move beyond antibiotics and really just see a much broader palate of options," Chase Beisel, leader of the RNA synthetic biology research group at the Helmholtz Institute for RNA-based Infection Research in Germany, told Live Science.
But until these new therapeutics are ready for prime time, the world needs to curtail its overuse and misuse of antibiotics, which experts say is speeding up the rate at which these lifesaving drugs become obsolete.
How antibiotic resistance emerges and spreads
Antibiotics either directly kill bacteria or slow their growth, leaving the immune system to finish the job. The drugs work in several ways — by preventing bacteria from building sturdy walls or making copies of their DNA, for instance. Growth-slowing antibiotics usually disrupt ribosomes, the factories in which bacterial cells make proteins.
Many antibiotics shoot for the exact same molecular targets, and so-called broad-spectrum antibiotics' mechanisms are so universal that they work on both major classes of bacteria: gram-positive and gram-negative, which are distinguished by the makeup and thickness of their cell walls. Broad-spectrum antibiotics, in particular, pressure both harmful and helpful bacteria in the body to evolve defensive strategies that eject or disable the drugs, or else alter their targets.
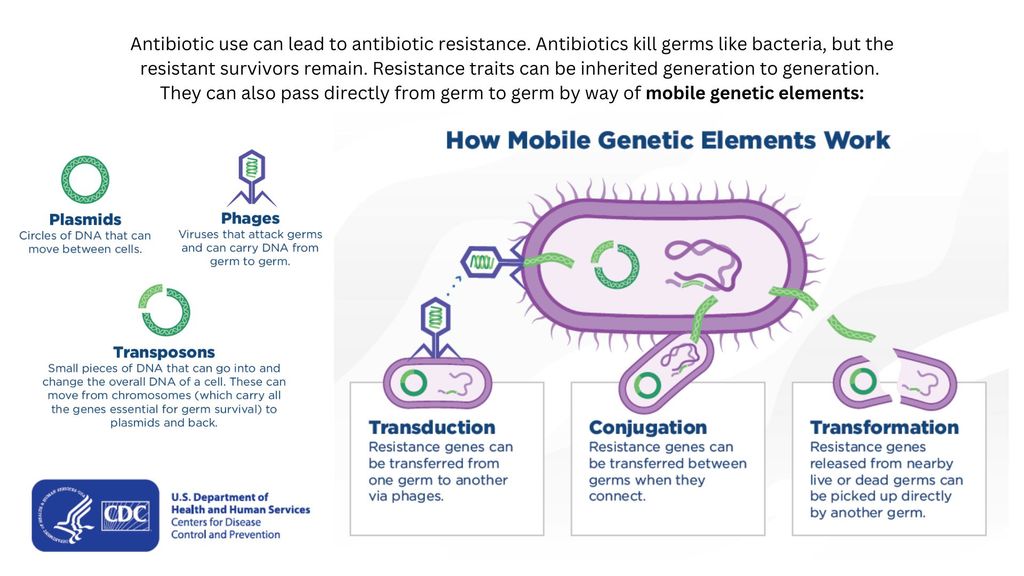
Drug-resistant bacteria can transfer their resistance to additional bacteria in several ways. (Image credit: Centers for Disease Control and Prevention. Adapted by Live Science from the CDC's "How Resistance Moves Directly Germ to Germ" Fact Sheet.)
Bacteria can pick up such defenses through random DNA mutations, or by swapping "resistance genes" with other bacteria via a process called horizontal gene transfer. By making these gene transfers, bacteria can quickly spread such mutations to additional bacterial populations in the body and in the environment.
The misuse of antibiotics in health care, as well as in agriculture, has given bacteria endless opportunities to develop resistance, raising the chance that once-treatable infections will become life-threatening.
Harnessing viruses to fight bacteria
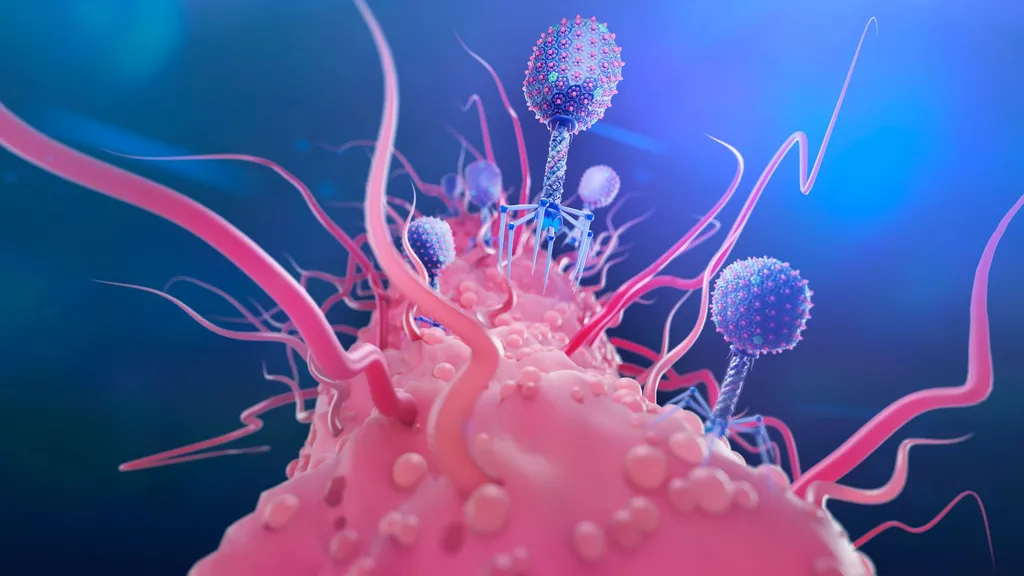
One of the proposed alternatives to antibiotics was first conceived more than a century ago, before the 1928 discovery of penicillin. Called phage therapy, it uses bacteria-infecting viruses called bacteriophages, or simply "phages," which typically kill the germs by invading their cells and splitting them open from the inside.
Phages can also pressure bacteria into giving up key tools in their drug resistance tool kits. For example, a phage called U136B can have this effect on E. coli. To infiltrate E. coli, the phage uses an efflux pump, a protein E. coli normally uses to pump antibiotics out of the cell. If the E. coli tries to change this pump to escape the phage, it reduces the bacterium's ability to pump out antibiotics.
And unlike with antibiotics, bacteria are unlikely to gain widespread resistance to phage therapy, said Paul Turner, director of the Center for Phage Biology and Therapy at Yale University.
Turner and other experts have concluded that, "if phage therapy were used at a global scale, that it would not lead to the same problem of widespread resistance to it, the way that antibiotic use has led to that problem," he told Live Science.
Here's why: Antibiotic resistance has been dramatically accelerated by the misuse and overuse of antibiotics, especially broad-spectrum antibiotics that work on a variety of bacteria. Phages, by contrast, can have much narrower targets than even narrow-spectrum antibiotics — for instance, targeting a protein found in only one or a few strains within one bacterial species.
The target bacterium can still evolve resistance to an individual phage — but by picking the right combination of phages, scientists can make it so that the bacterium's evolution comes at a cost, Turner said. This cost might be a decrease in virulence or an increased vulnerability to antibiotics.
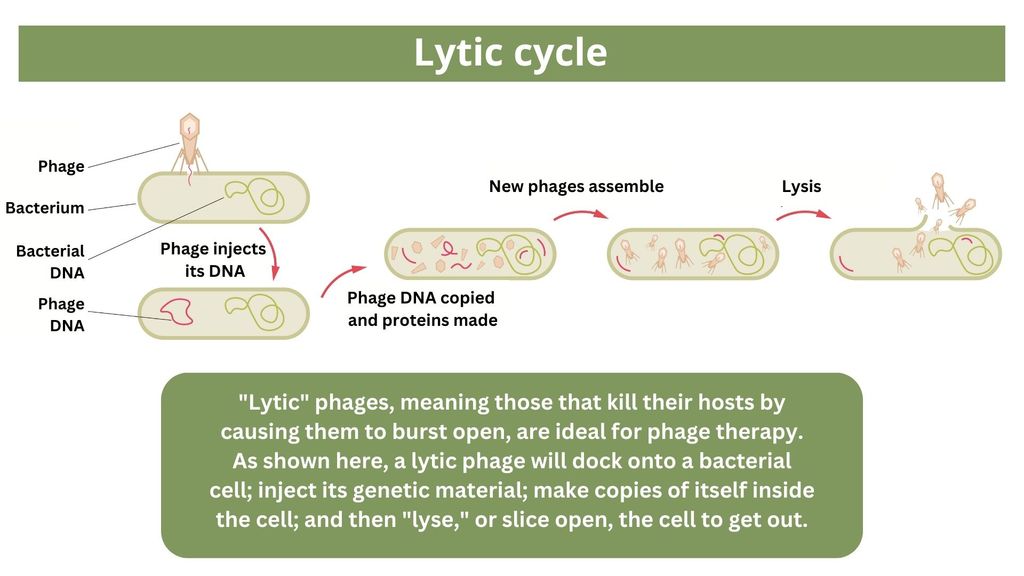
(Image credit: Graphic made by Olha Pohrebniak via Getty Images. Adapted by Live Science.)
To date, phage therapy has mostly been tested through a regulatory framework known as "compassionate use" in patients like the Brussels Airport bombing victim, whose infections had no other treatment options. Phage therapy has shown success in these settings, and in a recent observational study of 100 patients who received phages alongside antibiotics.
So far in clinical trials, though, phage therapy generally hasn't worked better than standard antibiotics or a placebo. Topline results from two recent trials hint at the treatment's effectiveness in specific lung and foot infections, but the full results have yet to be released.
Success in future trials will be key to getting phages into the clinic, Turner said. Those trials will have to show the therapy works for multiple types of infections, determine dosage and confirm phage therapies don't hurt helpful bacteria in the body, he added.
Turning bacteria's defenses against them
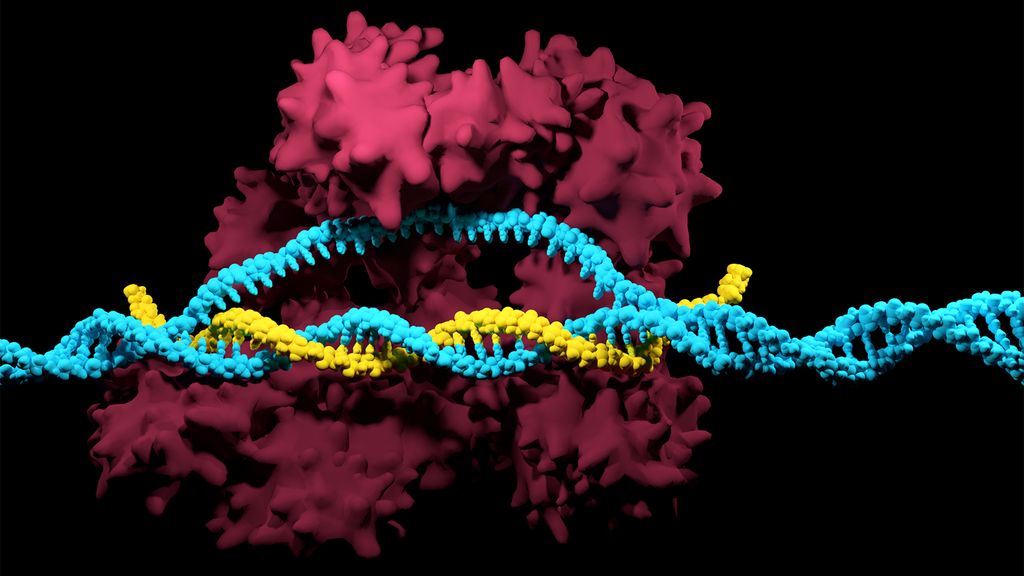
The CRISPR-Cas system can be used to snip DNA at precise locations. Here, a Cas enzyme (dark pink) is preparing to cut through a target DNA strand (blue) and is being told where to cut by an RNA strand (yellow). (Image credit: Meletios Verras via Getty Images)
Although made famous as a powerful gene-editing tool, CRISPR technology was actually adapted from an immune system found in many bacteria: CRISPR-Cas.
The key components of this immune system include molecular scissors, known as Cas proteins, and a memory bank of DNA snippets that a bacterium has collected from phages that once infected it. By tapping its memory bank, CRISPR-Cas can guide its lethal scissors to a precise point in an invading phage's DNA and snip it like a piece of ribbon.
On occasion, though, rather than attacking phages, CRISPR-Cas can accidentally go after the bacterial cell's own DNA, triggering a lethal autoimmune reaction. This phenomenon inspired Beisel and his colleagues to explore using CRISPR-Cas to shred bacterial cells' DNA.
"The real draw of it is that it is a sequence-specific tool," meaning it targets only the DNA you tell it to, and not sequences present in other bacteria, Beisel told Live Science. So, once administered to a patient, "the CRISPR machinery gets into a set of cells, but only those that have the sequence or sequences you picked will be attacked and killed."
How do you get CRISPR-Cas into the right bacteria? Various research groups are testing different delivery methods, but at present, the best strategy seems to be loading CRISPR machinery into a phage that infects the target bacterium, Beisel said.
Beisel is a co-founder and scientific adviser of Locus Biosciences, a biotech company that's currently testing a CRISPR-enhanced phage therapy in a midstage, roughly 800-person trial. This approach couples the bacteria-killing prowess of phages with the ability of CRISPR-Cas to destroy essential bacterial genes. As with CRISPR-less phage therapies, clinical trials are needed to determine the treatment's safety profile and appropriate dosing.
"I can see these [treatments] coming about in the five- to 10-year time frame," Beisel said.
Designer molecules to kill bacteria
Beyond phages and CRISPR, scientists are developing antibiotic alternatives that harness bacteria-slaying peptides — short chains of protein building blocks— and enzymes, specialized proteins that jump-start chemical reactions. These molecules differ from antibiotics because they can kill a very narrow range of bacteria by targeting bacterial proteins that cannot easily gain resistance to their attacks.
Lab-made molecules called peptide nucleic acids (PNAs) are some of the most promising candidates. These engineered molecules can be designed to block bacterial cells from building essential proteins that are crucial to their survival. PNAs do this by latching onto specific mRNA, genetic molecules that carry the instructions for building proteins from the cell's control center to its protein construction sites. PNAs cannot enter bacterial cells on their own, though, so they're typically attached to other peptides that easily pass through the bacterial cell wall.
By targeting proteins that cells cannot change without harming themselves, PNAs can avoid triggering drug resistance, Beisel explained. The engineered molecules could also be made to target proteins that directly contribute to antibiotic resistance, for example, the efflux pumps used to push antibiotics out of cells or the enzymes capable of disabling the drugs. By emptying a germ's drug resistance tool kit, PNAs can then make it vulnerable to standard treatments.
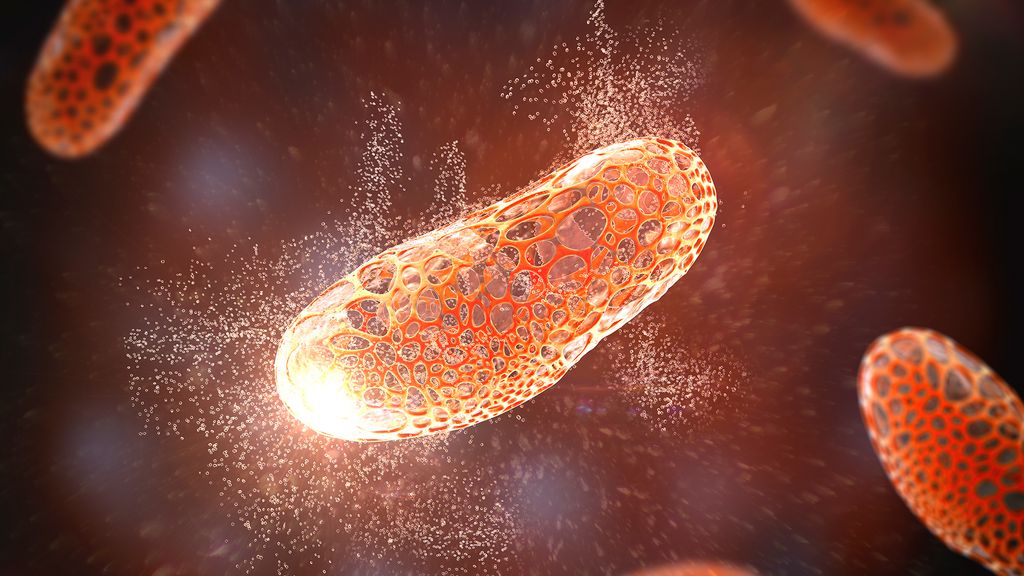
One approach for killing bacteria is to use lysins, or enzymes that tear open bacterial cell membranes and cause the microbes' contents to spill out. (Image credit: KATERYNA KON/SCIENCE PHOTO LIBRARY via Getty Images)
Antibacterial PNAs are still being tested in lab dishes and animals and have not yet moved into human trials. And, scientists need to make sure PNA-based treatments don't inadvertently mess with human cells or helpful bacteria.
In addition to peptides like PNAs, enzymes called lysins are another promising treatment option. Lysins are used in nature by phages to split bacteria open from the inside. They act like tiny swords that slice through the outer wall of a bacterial cell, spilling its guts. The molecular sabers are unlikely to promote resistance because bacteria cannot easily change the essential cell-wall components that lysins target.
Lysins slaughter bacteria quickly upon contact, and they can be very specific, killing some types of bacteria while sparing others. Furthermore, lysins can be tweaked in the lab to change which bacteria they target, boost their potency and improve their durability in the body.
Some lysins have entered mid- and late-stage human trials with hundreds of participants, in which they've been tested as supplementary treatments to antibiotics but garnered mixed results.
Antibiotic stewardship can save lives, in the meantime
Until these next-gen bacteria slayers make it to market, immediate measures must be taken to stall the rise of superbugs, by preventing the misuse of antibiotics that pressures bacteria to evolve resistance in the first place.
For example, doctors can be more diligent about confirming that bacteria, not viruses, are behind a patient's infection before prescribing antibiotics, said Dr. Shruti Gohil, a lead investigator of four INSPIRE-ASP Trials, federally funded research aimed at improving hospitals' antibiotic use. Other safeguards can include auditing doctors' prescriptions to see if narrower-spectrum drugs could be used instead of broad ones, or requiring special clearance for the broadest-spectrum drugs. These steps are essential not only in hospitals but everywhere antibiotics are prescribed, from primary care to dentistry, Gohil said.
Each interaction between a doctor and their patient matters.
Gohil stressed that "by reducing individual risk, you anticipate that you will drop the overall population-level risk," and eventually slash the prevalence of multidrug-resistant bugs.
- Karlston
-

 1
1


Recommended Comments
There are no comments to display.
Join the conversation
You can post now and register later. If you have an account, sign in now to post with your account.
Note: Your post will require moderator approval before it will be visible.